intro-to-galaxy-ngs-sarscov2
Introduction to Galaxy
Galaxy is a web-based platform for running data analysis and integration, geared towards bioinformatics.
- Open-source, public servers
- Developed at Penn State, Johns Hopkins, OHSU and Cleveland Clinic with many more outside contributions
- Large and extremely responsive community
Log in to Galaxy and open workshop repository
-
Connect to Tufts Network, either on campus or via VPN
- Using a Chrome browser, visit https://galaxy.cluster.tufts.edu
- Note: sometimes an incognito browser is needed
-
Log in with your Tufts credentials
-
In another browser window go to course website: https://rbatorsky.github.io/intro-to-galaxy-ngs-sarscov2
- If you have one monitor, here is a suggested screen layout to follow the tutorial
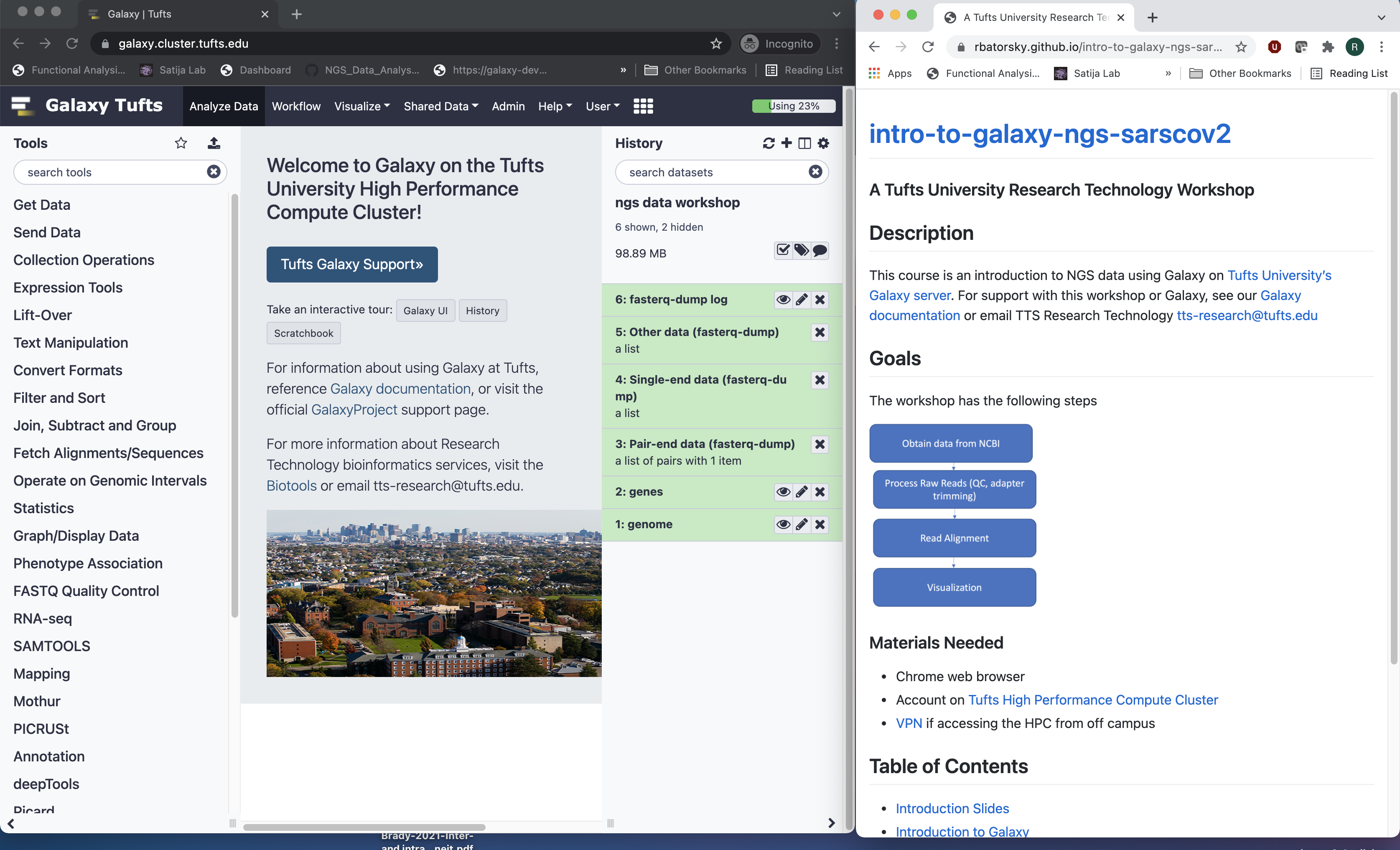 Suggested screen layout
Suggested screen layout
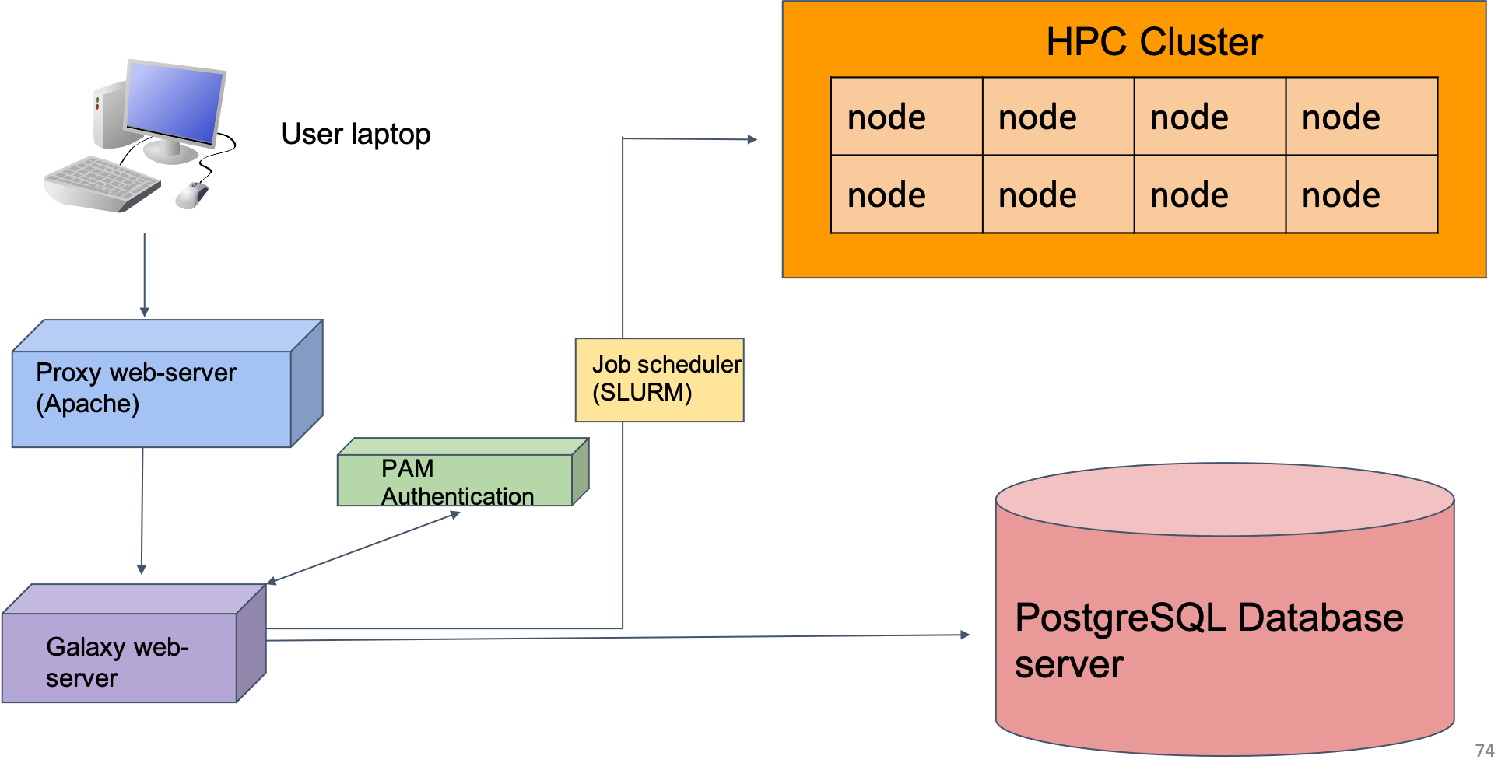
Galaxy on the Tufts University High Performance Compute (HPC) Cluster
- Our Galaxy server runs on the HPC cluster, storing data on HPC drives and running user jobs on compute nodes
- In practice, this means that Tufts users have more resources on Tufts’ Galaxy compared to public Galaxyservers
- We also have the ability to install and configure open source and custom tools
- More information on getting access to Tufts Galaxy can be found on the Research Technology website
Galaxy UI
- We’ll spend some time getting familiar with the Galaxy interface, layout, and key functions
- The Galaxy User Interface has a top menu bar and three panels: Tools, Main, and History
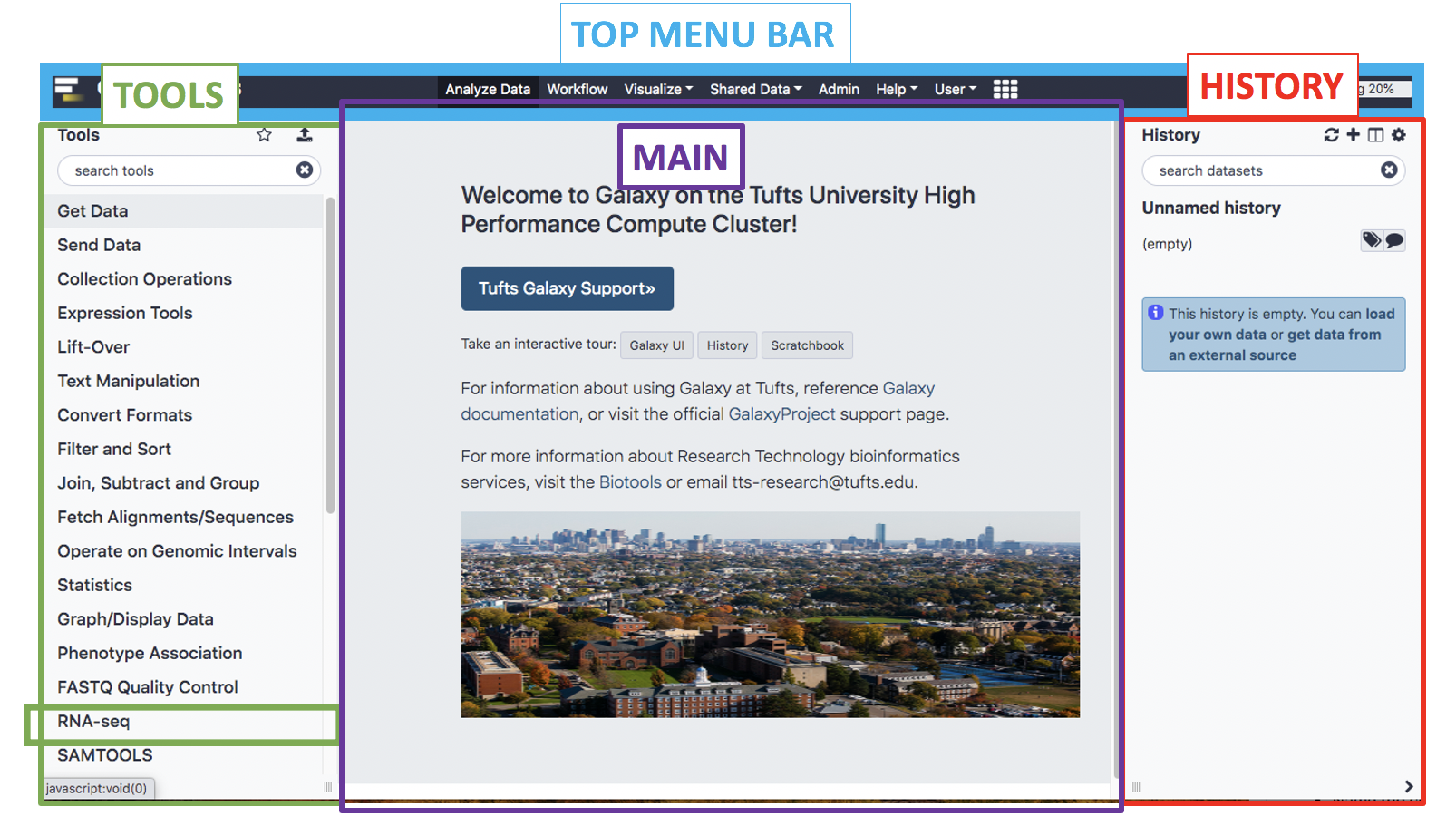
- The panels can be expanded/minimized using the small arrows and dragging the three small lines in the bottom left and right corners of the page
- To get back to the home page click the logo next to Galaxy Tufts on the top left of the page or the Analyze Data tab on the top of the page
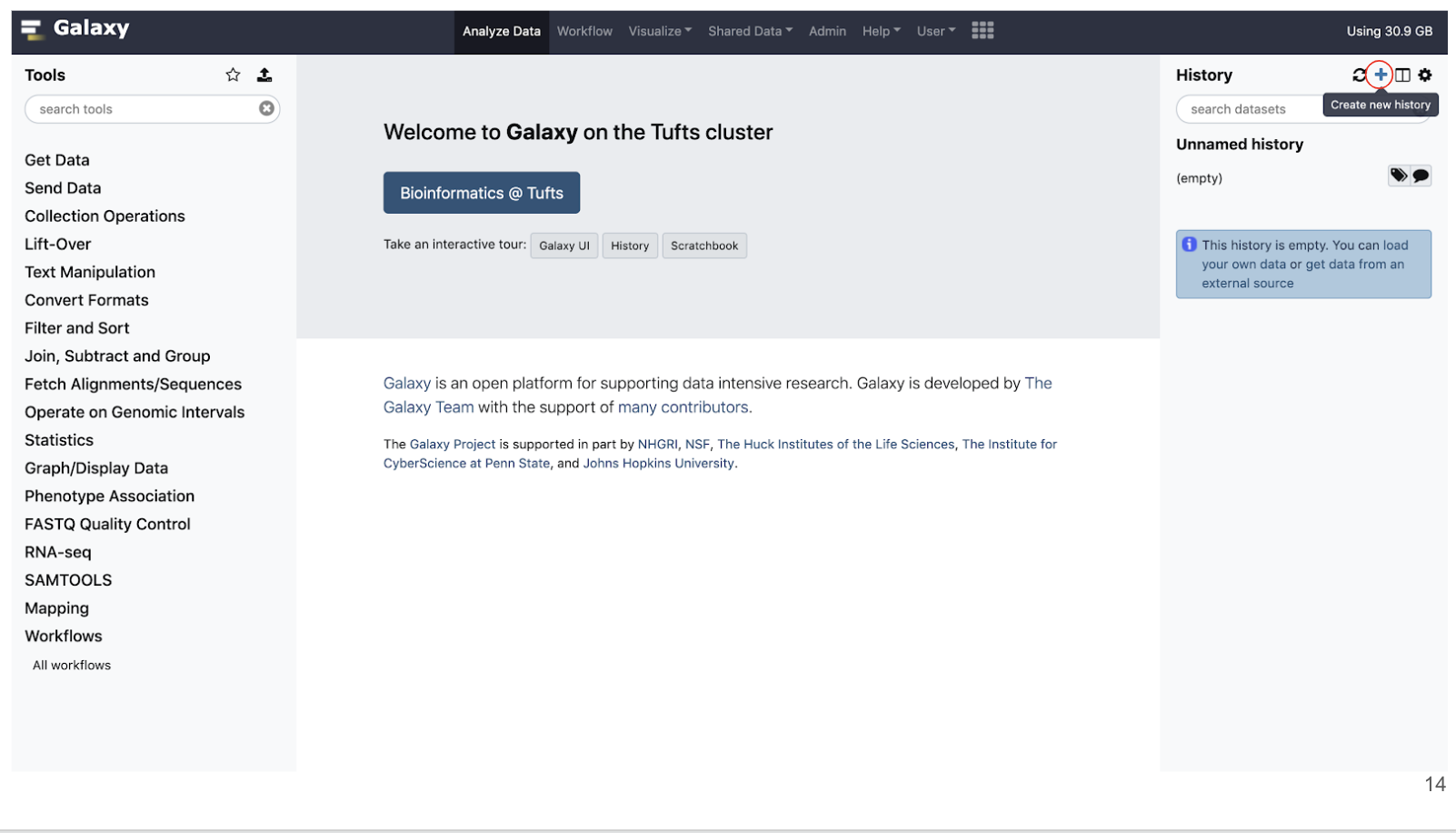
Histories
- Histories are where a dataset and a set of analysis are stored
- Similar to a “working directory”.
- You can have multiple histories under the same account and you can create a new one by clicking the + as shown below
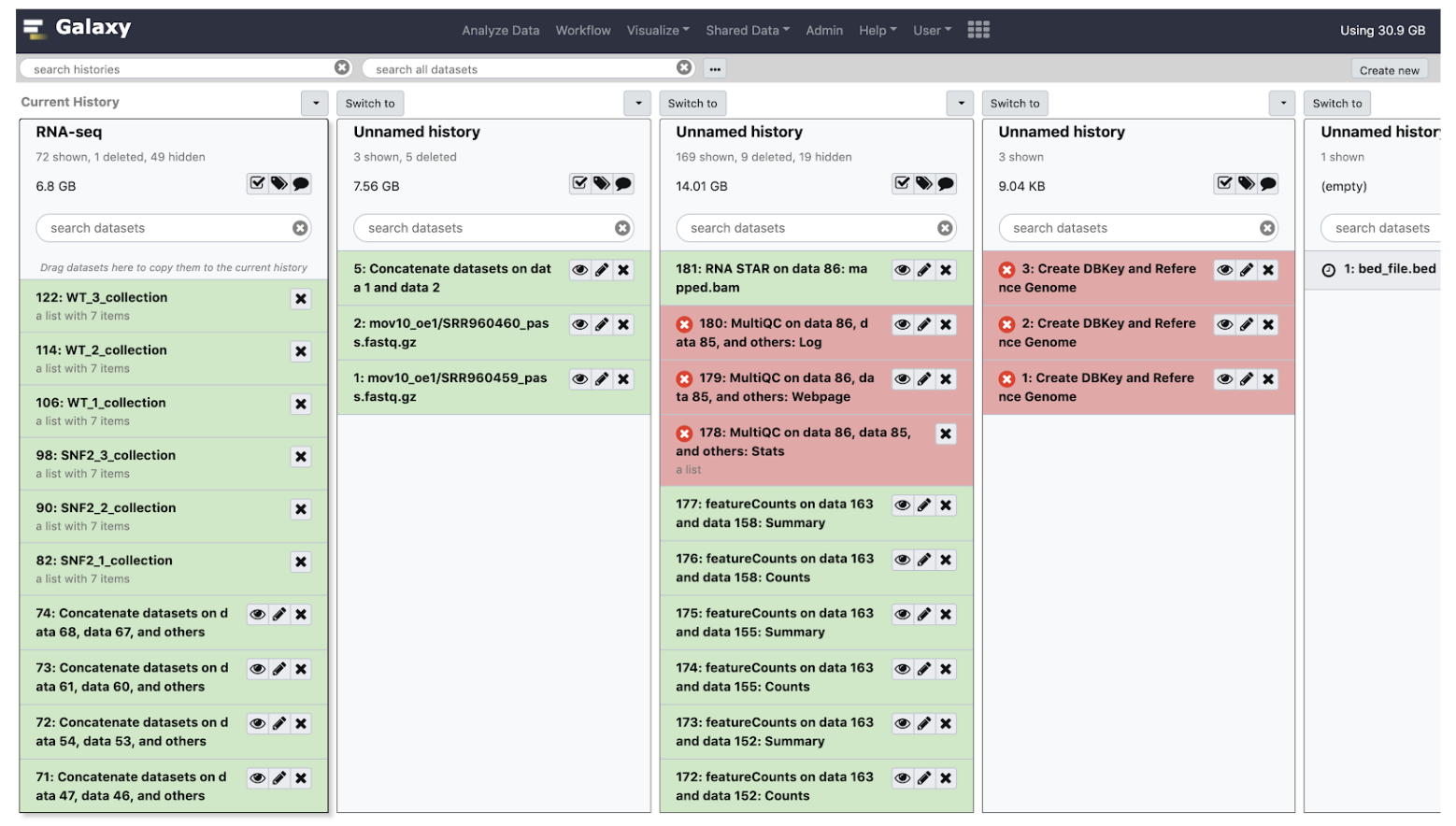
- Here you can view all your histories, switch to another history, and drag and drop data between histories.
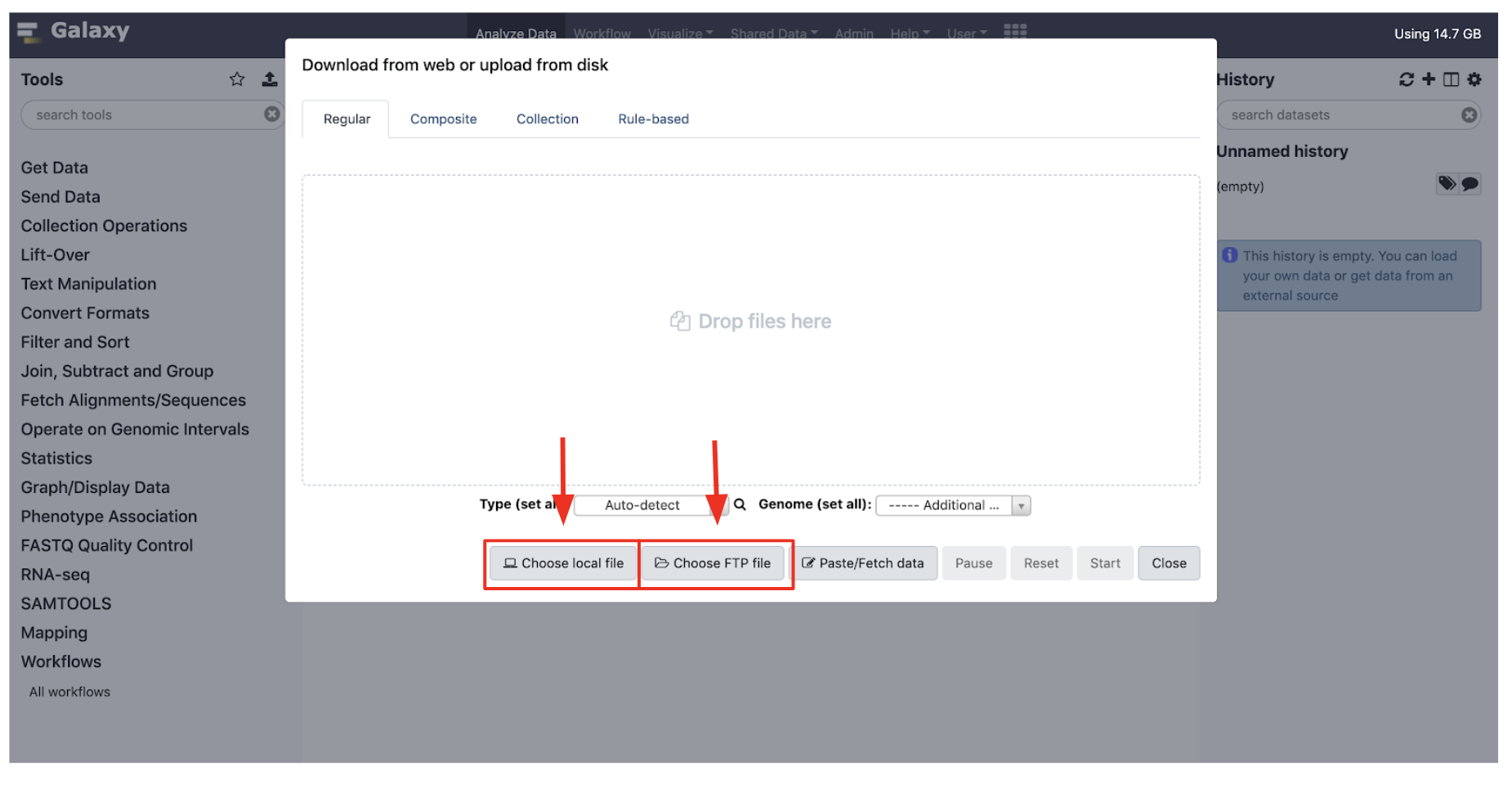
Uploading Data
- Data can be uploaded to Galaxy in a number of ways by clisking the “Upload” icon on the top of the Tools panel.
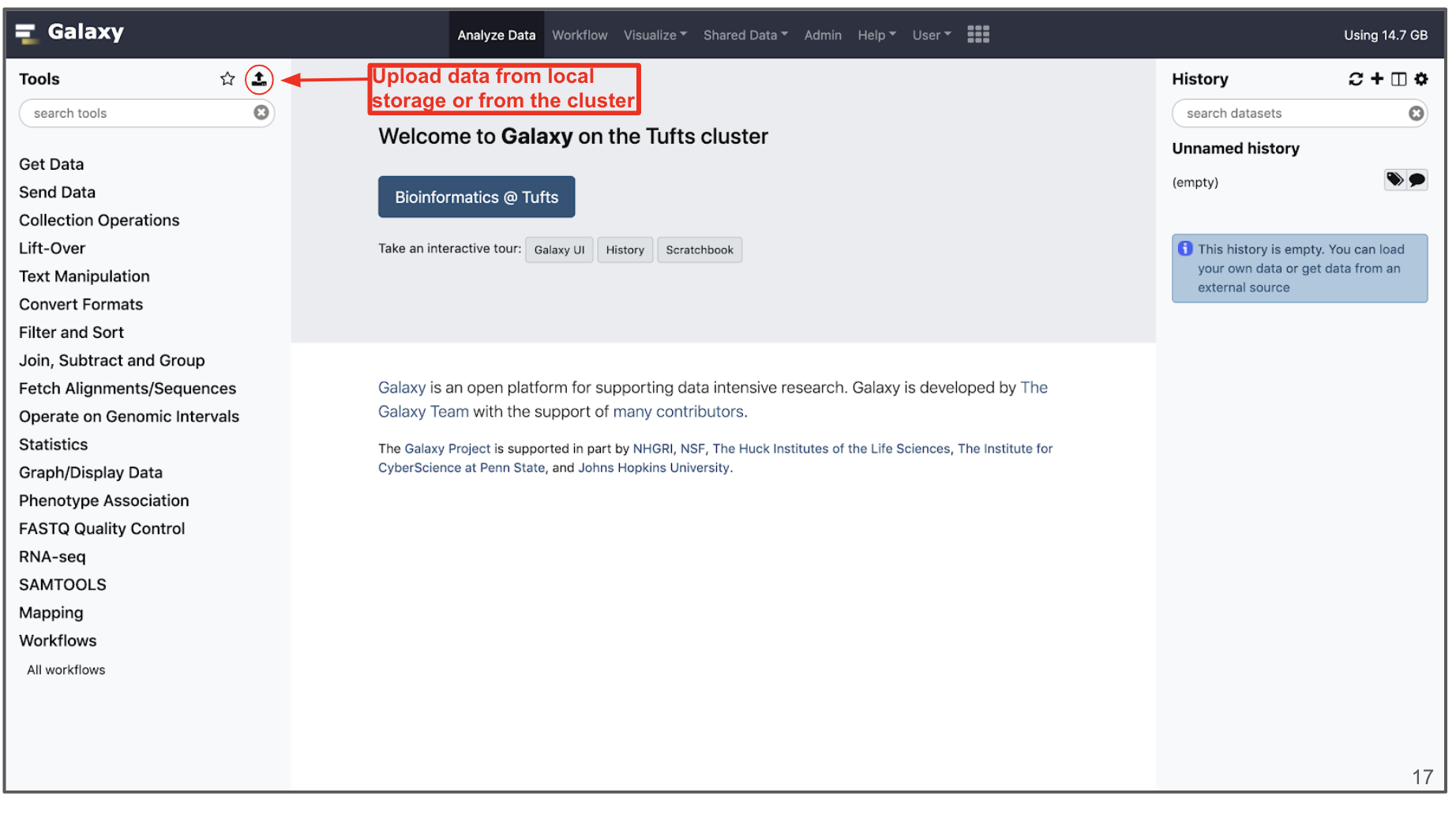
- “Choose local File” will select a file from your local computer, and “Choose FTP File” will select from your user drive on the HPC cluster, which has an extension “/cluster/tufts/galaxy/xfer/username”
Tools
- On the Tools panel, either search by name or click the category that your tool would fall under
- Our tools for today will mostly be stored under the RNA-seq category
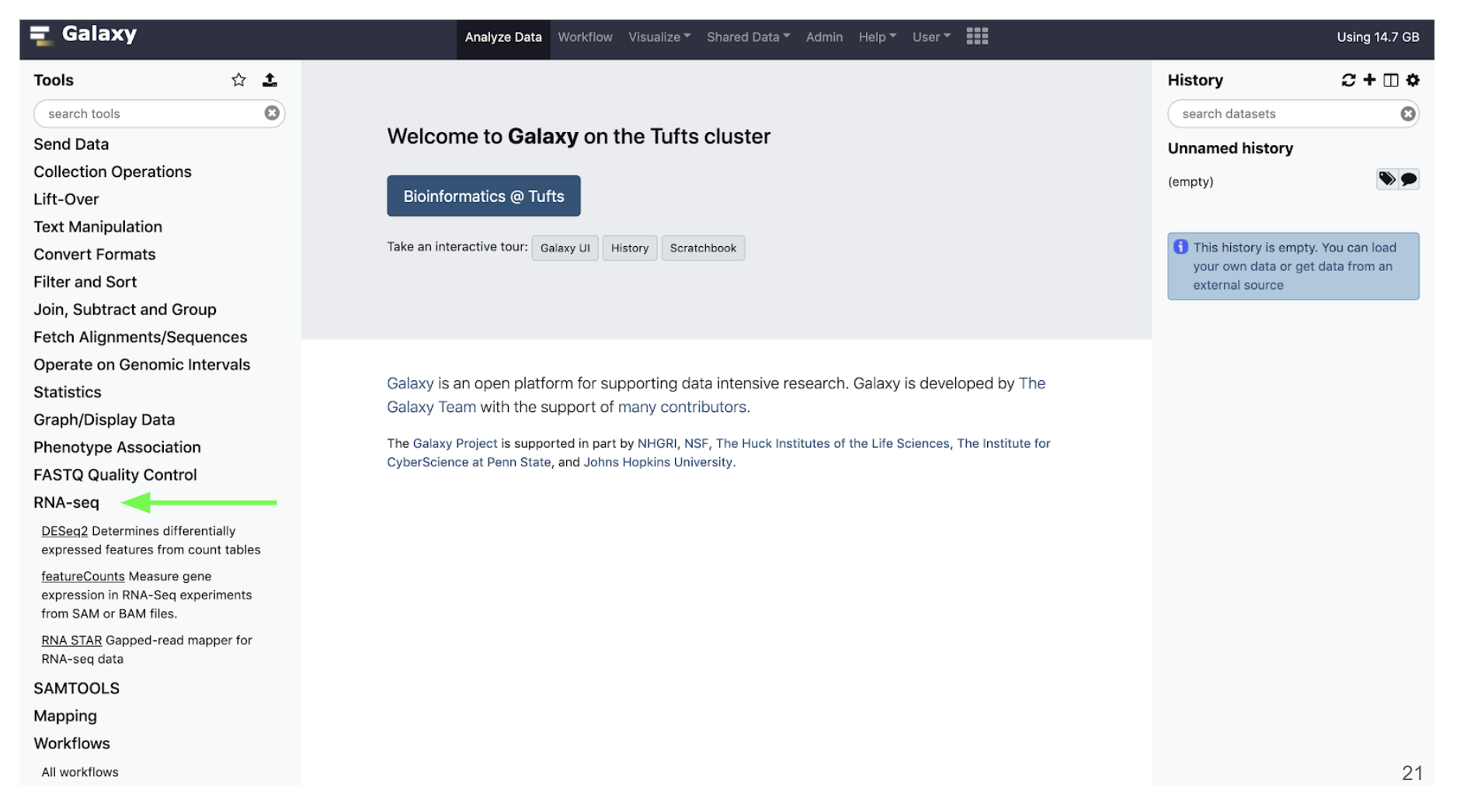
- Click the name of the tool you would like to use and the tool menu will appear in the Main panel