intro-to-rnaseq-with-galaxy
Perform Quality Control on Raw Reads
Introduction to FastQC
FastQC provides several modules (as discussed in intro Slides)
- Sequence Quality
- GC content
- Per base sequence content
- Adapters in Sequence
Run FastQC
- In the Tools panel search bar, type FastQC
- Select FastQC under FASTQ Quality Control (You may need to click Show Sections to see section headers)
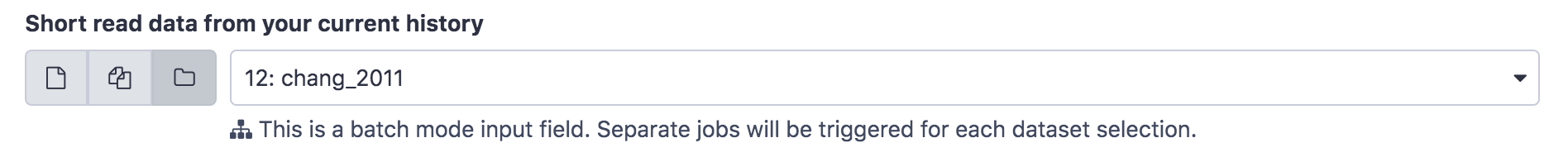
- In the Main panel, under Raw read data from your current history select the folder icon
 and the dataset subsampled_chang_2011 will appear as an option
and the dataset subsampled_chang_2011 will appear as an option
- Scroll down and click Execute.The job should first appear orange and then green after a minute or so.
- The result will be two lists, one containing the raw data and one the webpage (html) results for convenient viewing in the browser.
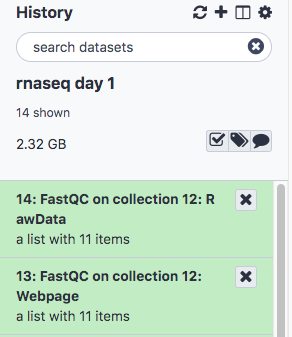
- Click to expand the second list FastQC on collection 12: Webpage and click on the
 next to the first file for sample HIV_12hr_rep1. The first table gives Basic Statistics of the sample. The Main panel will show metrics and plots. You may have to adjust the size of the panel in order to view.
next to the first file for sample HIV_12hr_rep1. The first table gives Basic Statistics of the sample. The Main panel will show metrics and plots. You may have to adjust the size of the panel in order to view.
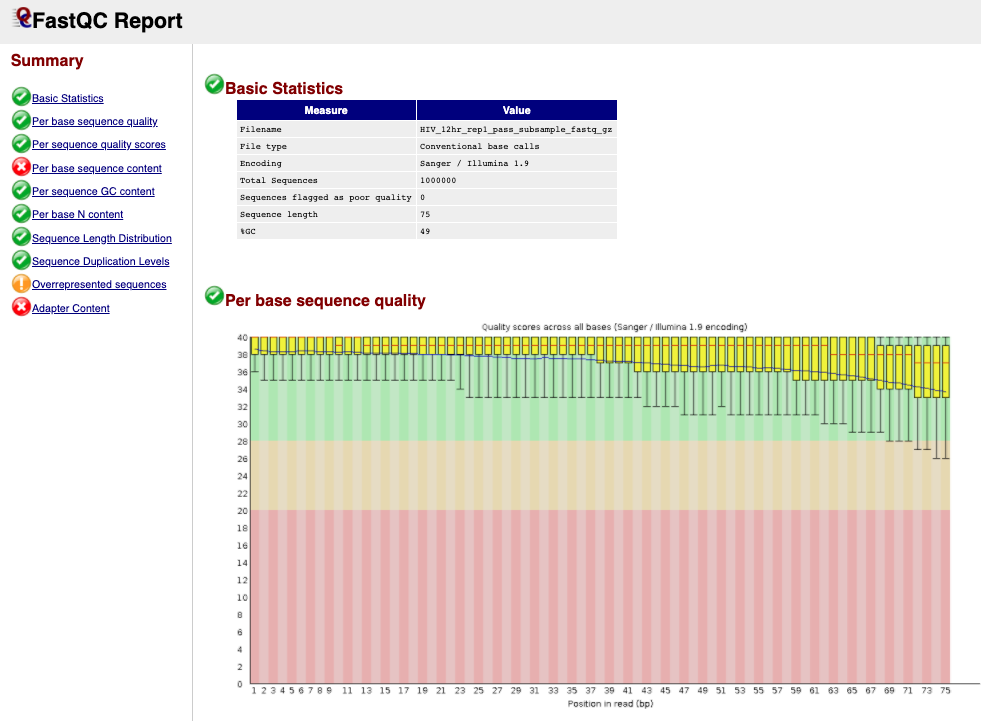
Question 2: Were you right about your guess of quality encoding?
Aggregate QC data with MultiQC
The tool MultiQC allows us to view our QC results from all samples side by sides, in order to check for consistency across replicates. It can use the Raw Data output from FastQC and generate plots for all modules.
Steps to run:
- In the Tools panel search bar, type MultiQC
- Select MultiQC under FASTQ Quality Control
- In the middle panel, under Which tool was used generate logs? select FastQC
- Under FastQC output click the
 and select the collection 14: FastQC on collection 12: Raw Data (note that the numbers 14 and 12 are tracking the dataset number in your history and might vary if you have not followed the exact sequence in this document)
and select the collection 14: FastQC on collection 12: Raw Data (note that the numbers 14 and 12 are tracking the dataset number in your history and might vary if you have not followed the exact sequence in this document) - Enter the Report Title “Raw data QC”
- Scroll down and click Execute.
- The result will again be two collections (you may have to click “back to rnaseq day 1” on the top of the History panel). Select the collection titled MultiQC on data 36, data 34, and others: Webpage and click the
 to view. (If panels show “loading” for more than a few seconds, click the
to view. (If panels show “loading” for more than a few seconds, click the  a second time to refresh)
a second time to refresh)
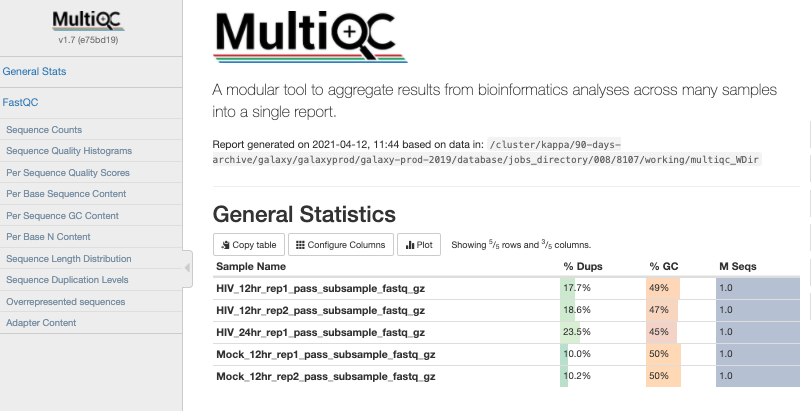
Question 3: Using what we learned in lecture, which metrics show one or more failed samples?
Trim adapters and low quality read ends with Trim Galore!
- In the Tools panel search bar, type Trim Galore!
- Select Trim Galore! under FASTQ Quality Control
- Under Reads in FASTQ format click the
 and select chang_2011
and select chang_2011 - Scroll down and click Execute.
- The result will be a single collection titled Trim Galore! on collection 12: trimmed reads. Next, we’ll rerun FastQC in order to see how the trimming performed
Rerun FastQC and MultiQC
- Follow the steps for Run FastQC and Aggregate QC data with MultiQC above, except select the trimmed reads generated in the previous step as the input to FastQC